Compute a temporal aggregation and plot on a map
Import PyOphidia and connect to server instance
[1]:
from PyOphidia import cube, client
cube.Cube.setclient(read_env=True)
Current cdd is /home/ecasuser
Current session is https://ophidialab.cmcc.it/ophidia/sessions/28428206836738622831574257875438385/experiment
Current cwd is /
The last produced cube is https://ophidialab.cmcc.it/ophidia/6215/849435
Import data, compute average over time and extract values
[2]:
mycube = cube.Cube.importnc(src_path='/public/data/ecas_training/tos_O1_2001-2002.nc',measure='tos',imp_dim='time',import_metadata='yes',ncores=5)
mycube2 = mycube.reduce(operation='max',ncores=5)
mycube3 = mycube2.rollup(ncores=5)
data = mycube3.export_array()
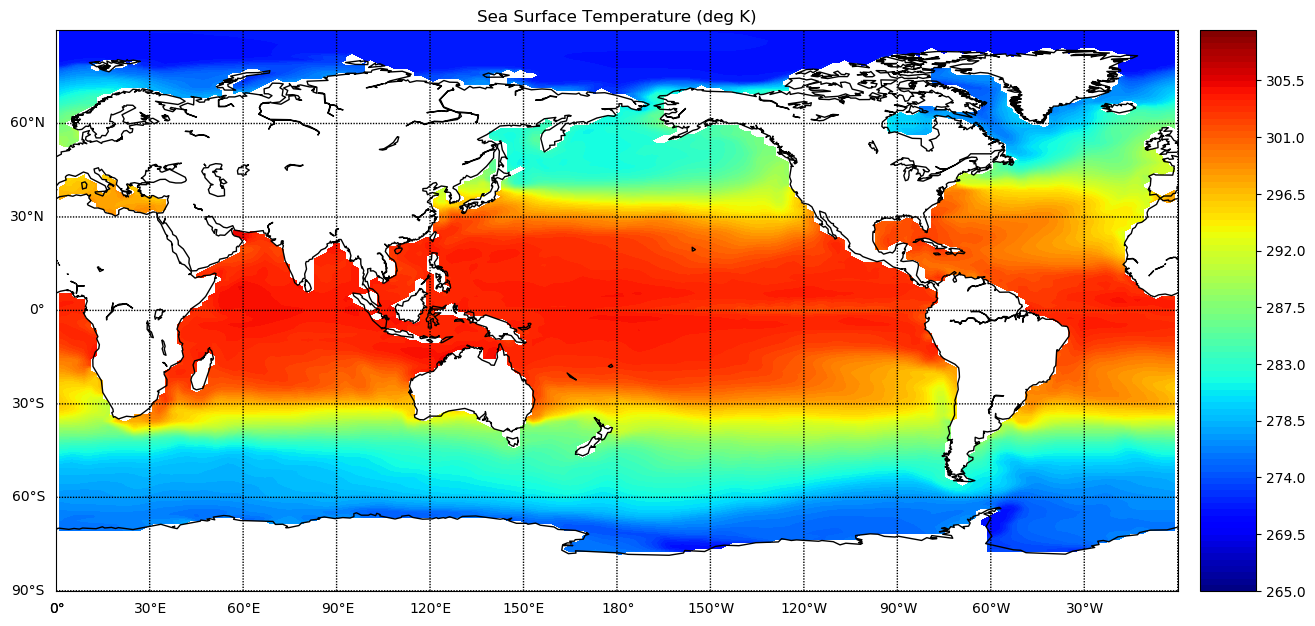
Plot output data on map
[3]:
%matplotlib inline
import matplotlib.pyplot as plt
from mpl_toolkits.basemap import Basemap, cm
import numpy as np
lat = data['dimension'][0]['values'][:]
lon = data['dimension'][1]['values'][:]
var = data['measure'][0]['values'][:]
fig = plt.figure(figsize=(15, 15), dpi=100)
ax = fig.add_axes([0.1,0.1,0.8,0.8])
map = Basemap(projection='cyl',llcrnrlat= -90,urcrnrlat= 90, llcrnrlon=0,urcrnrlon=360, resolution='c')
map.drawcoastlines()
map.drawparallels(np.arange( -90, 90,30),labels=[1,0,0,0])
map.drawmeridians(np.arange(-180,180,30),labels=[0,0,0,1])
x, y = map(*np.meshgrid(lon,lat))
clevs = np.arange(265,310,0.5)
cnplot = map.contourf(x,y,var,clevs,cmap=plt.cm.jet)
cbar = map.colorbar(cnplot,location='right')
plt.title('Sea Surface Temperature (deg K)')
plt.show()
Export result to NetCDF file
[4]:
mycube3.exportnc2(output_path='/home/' + cube.Cube.client.username,export_metadata='yes')
A file called tos.nc will be created in your home folder
Clear your workspace before running other notebooks
[5]:
cube.Cube.deletecontainer(container='tos_O1_2001-2002.nc',force='yes')